by Phahlamohlaka Mokgohlwer Cynthia
Due to numerous limitations that come with the study and clinical application of the genetic engineering technique CRISPR on tumor cells. This research addresses some of the important and crucial factors, accuracy, and robustness cautiously on cancer therapy. Incorrect genetic editing and modification in the medical domain can result in severe consequences and cause adverse outcomes (more diagnoses). Here individual cells are strategically targeted for sgRNA integration in pooled library CRISPR screening, with careful reduction of cell usage.
After a limited number of genes were screened. A new improved method was established, the genome-scale CRISPR screening of TGF-beta tumor suppressor pathways in human small intestinal(hSI) organoids. Tumor suppressor genes inhibit all the genes that promote cancer cell progression. It has been found that cancer originating from intestinal stem cells mutation often involves various signaling pathways. APC gene is responsible for Colorectal cancer through activation of the Wnt pathway. The progression of the cancer cells to spread beyond their original site requires the accumulation of additional tumor driver mutations. To prompt the intestinal stem cell differentiation, cell-autonomous (signal transduction) inhibition of the pathway through transforming growth factor-beta (TGF-beta) inactivation, which enables cancer cells tolerance to TGF-beta concentration. Gradually the tumor becomes independent of the signal niche.
This medical research required the use of Organoids, imitating the human small intestinal stem cells and colon cells, discovered as the closest model of real gastrointestinal tissues, with the respective signaling pathways. Both wild-type (WT) and APC mutants were used to precisely and inclusively identify the genes responsible for TGF-beta resistance. The healthy and wild donors were cultured under the same conditions in a 3D organoid system as organoids are an alternative for living cell lines in Genome-wide CRISPR screening, better immobilized for cell accessibility and analysis. The targeted genes in the organoids are transduced with the lentiviral library for knockout. The procedure took place co-currently for Wnt pathway and TGF-beta pathway under selective conditions on human small intestine organoids(hSI) and colorectal cancer organoids (CRC) respectively.
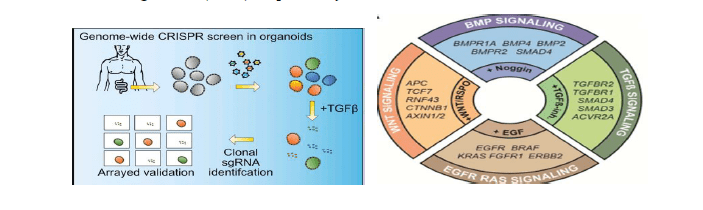
The efficient transduction of lentiviral library (1, 698 sgRNA) and survival of human small intestine organoids, for successful evaluation of the negative Wnt-regulators in Wnt pathway by targeting 283 potential tumor suppressor genes. The sgRNA as a director gene will direct the gene editor/scissor-like (CRISPR) to bind to the correct targeted gene, modifying the gene function. Incubated each cell with 4 different sgRNAs/CRISPR library and screened individually to avoid false positive results, in 15 days 385 of 19 114 common cancer cell lines were identified. Based on the Wnt pathway screening (selection condition) only the outgrowing organoids were isolated and amplified, and the individual clones were assigned sgRNA using deep sequencing. To discern clonally integrated sgRNA reads from the background reads, for easy analysis as shown below.

Figure 1: CRISPR Screening in Organoids with Pooled sgRNA Libraries. Experimental outline for lentiviral organoid transduction (upper left), traditional pooled CRISPR screening (lower left), and pooled CRISPR screening with single organoid analysis (right).
Colorectal cancer (CRC) organoids were used to investigate the resistance of TGF-beta core pathway components for inhibition of TGF-b-mediated growth. TGF-beta selection assay involved TGF-beta inhibitors replacement with TGF-beta1 ligand driving intestinal CRC organoids to apoptosis. Conditionally, a significant percentage of TGF-beta-resistant organoids contained sgRNAs targeting known TGF-b pathway components, compared to the 4% of the cell clones with unknown linkage to TGF-beta signaling. The APC mutant (APCKO) hSI organoids were further screened for TGF-beta resistance, which may rely on Synergistic interaction with APC mutant APCKO/hSI organoids. There was a major difference in transcriptional response of the TGF-beta pathway between APC and wild-type organoids. Only 19% of organoids transduced with tumor suppressor gene TP53, grew under TGF-beta selection and contained sgRNAs against TGF-beta receptors, which suggested less restrictive conditions for further identification. TGF-beta pathway depends on the tumor occurrence site and pathway involved, while Wnt depends on function loss mutations.
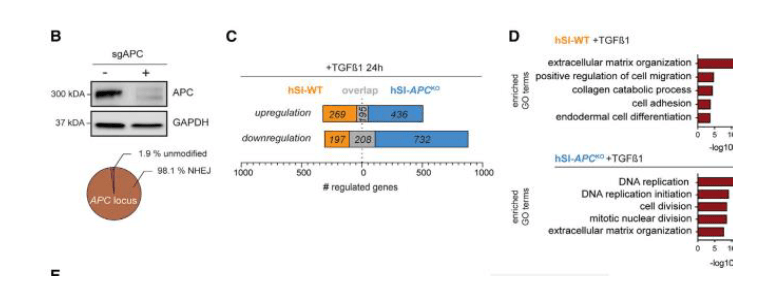
Figure 2: Single Organoid Comparison and Analysis of Genome-wide CRISPR-Cas9 Screening in Human Organoids.
Further introduced array rescreening to distinguish the functional hits of sgRNA integration with targeted TGF-beta components (TGFBR1, TGFBR2, SMAD3, and SMAD4) to attain their impact on TGF-beta resistance in WT-hSI organoids. Discovered APCKO organoids SWI/SNF components (ARID1A and SMARCA4) along with genes while WT background screening confirmed only FAM122A as a functional hit. The functional hits are a potential cause of resistance. Models induced resistance through combining WT organoids with mutant APC, associated with SWI/SNF components in TGF-beta treatment assay as shown below. The TP53KO-exposed non-transduced organoids outgrow in TGF-b selection conditions. The SWI/SNF complex component (ARID2) concludes resistance to TGF-b-mediated growth inhibition in mutated intestinal organoids. The accuracy and robustness of the single organoid after Traditional genome wide CRISPR screening identified only TGFBR2 as the APCKO organoids resistance factor.
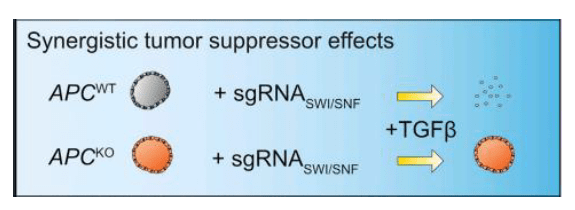

Figure 3: Validation of sgRNAs that Cause Resistance to TGF-b-Mediated Growth Inhibition in hSI Organoids.
Mutating ARID1A and SMARCA4 in human APCKO/P53KO/KRASG12V intestinal organoids, enhanced the formation of tumor modules after xenotransplantation. SWI/SNF mutant organoids show significant growth over SWI/SNF mutant organoids in TGF-beta-rich environment.
As SMAD4 is a phosphorylated stable form of the TGF-beta receptor, it is translocated to regulate target gene transcription in the nucleus. Mutations in ARID1A and SMARCA4 attenuate the signal transduction from the TGF-b receptor to the SMAD complex, recruited by cofactors, and promote tumorigenesis. To track the SMAD complex function, phosphorylation induction in APCKO/ARID1AKO and APCKO/ SMARCA4KO organoids was conducted and led to SMAD upstream signal transduction, as shown in Figure 4A. RNA-seq of different genotypes evaluated 443 of 1,571 expressed gene were dependent on ARID1A or SMARCA4. As such TGF-beta-SMAD-signaling cascade is still functional and translocates with reduced transcription response of TGF-b-target genes.

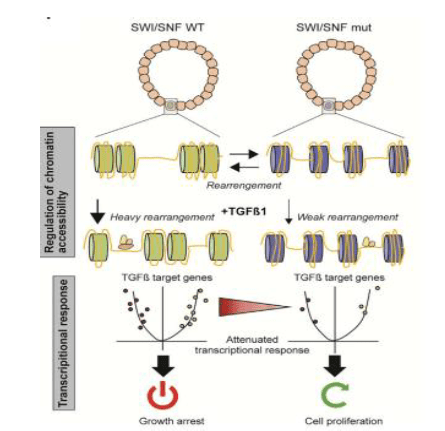
Figure 4: SWI/SNF (SMAD) regulation and Transcriptional response in Wild and APC mutant
CONCLUSION:
Successful screening was achieved for resistance mechanisms to the tumor-suppressive effects of TGF-b and identified several components of the SWI/SNF chromatin remodeling complex. SWI/SNF investigation shed light onto transcriptomic changes upon SWI/SNF perturbation which affect SMAD cofactors expression, thereby indirectly altering TGF-b target gene regulation and promoting cancer progression. The effectiveness and accuracy of the method gave concise and reproducible results from the stored libraries.
CITATION:
Ringel, T. et al. (2020) ‘Genome-Scale CRISPR Screening in Human Intestinal Organoids Identifies Drivers of TGF-β Resistance’, Cell Stem Cell, 26(3), pp. 431-440.e8. Available at: https://doi.org/10.1016/j.stem.2020.02.007.
Leave a comment